GC Image GCxGC Edition is a system designed for visualizing, processing, analyzing, and reporting on data produced in two-dimensional gas chromatography (GCxGC). GC Image GCxGC Edition has three main interfaces:
This introduction provides a brief overview of GCxGC data and a quick tour of some of the image processing facilities of GC Image GCxGC Edition. Chapter 2 contains installation instructions for the software. Chapters 3-17 describe the GC Image interface and chapters 18-20 describe GC Project. Chapters 21-22 contain instructions for calibration and reporting. Chapter 23 introduces the Help Resources for the software. Appendix A provides Answers to Frequently Asked Questions . Appendix B is a white paper on GCxGC Blob Metadata and Statistics in GC Image. Appendix C describes the plugin interface for the software.
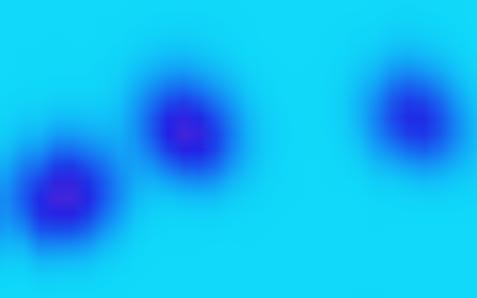
GCxGC separates chemicals according to the time each chemical requires to pass through two capillary columns. GCxGC data can be displayed as an image with pixels arranged so that the abscissa (X-axis) is the retention time for chemicals to pass through the first column and the ordinate (Y-axis) is the retention time for chemicals to pass through the second column. Each pixel value indicates the rate at which molecules are detected at a specific time.The elution of a compound(s) in the sample produces a small blob or cluster of data-point pixels with values that are larger than background values. The magnitudes of the data-point pixel values indicate the quantity of the chemical(s) present. fig.threeBlobs illustrates a greatly magnified view of a small region of a GCxGC image containing three blobs of pixels indicating (at least) three separated chemicals. The smaller values of the background are colorized light-blue and the larger values of the blobs are colorized dark-blue and magenta.
 |
| fig.threeBlobs: A region of a GCxGC image containing three blobs of pixels indicating three separated chemicals. |
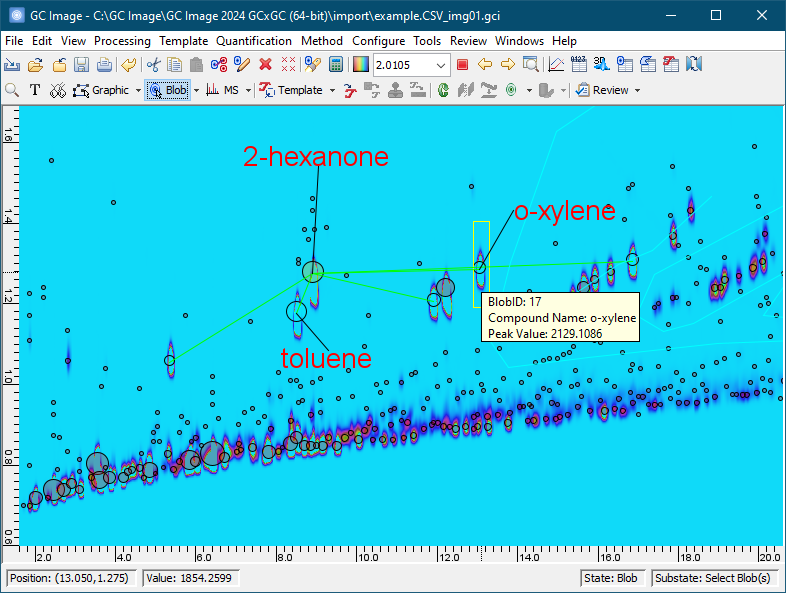
The position (i.e., the retention times in each dimension) of each blob is related to the physical properties the chemical(s) that produced the blob, so the position of a blob is useful in identifying each chemical in a sample. The sum of the pixel values in each blob is related to the quantity of the chemical that produced it, so the sum of pixel values of a blob is useful in quantifying the amount of each chemical in a sample.
GC Image has many tools that support various operations. This quick tour illustrates some of the steps in a typical processing sequence to analyze raw data produced by GCxGC, identifying and quantifying chemicals in a sample:
The following figures illustrate each of these steps. Later sections of the GC Image GCxGC Edition User Guide describe these and other operations in greater detail.
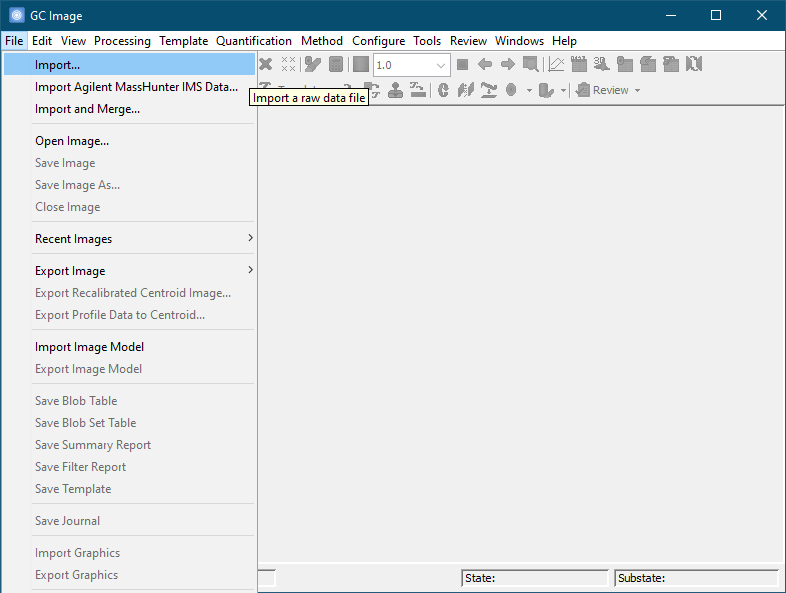
Locate the desired source data file and click the Open button. For example, a source file containing the raw chromatographic data can be a text file in comma-separated-value (CSV) format. The destination file name is automatically generated by GC Image, but can be changed by the user during the save operation.
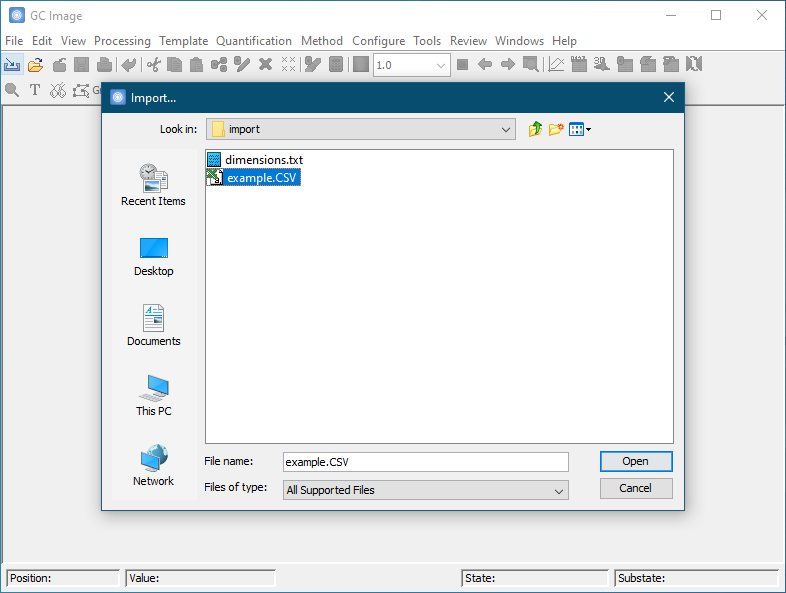
Specify the dimensions and optionally specify other processing options. The dimensions of the data are required and will be read from the source file if provided by the file format or may be specified by the user in pixel or time units. For example, the dimensions of data acquired at 200 Hz with a modulation period of 3 seconds and a run time of 33 minutes could be sized equivalently as 660 pixels for the first dimension (33 minutes / 3 seconds / modulation) and 600 pixels for the second dimension (200 samples/second * 3 seconds).
Specification of a configuration file and processing operations are optional in this pop-up. These optional specifications are a convenient mechanism for quickly directing processing at the time data is imported. Chapter Configurations describes these capabilities. The rest of this quick tour demonstrates how the processing operations are performed interactively.
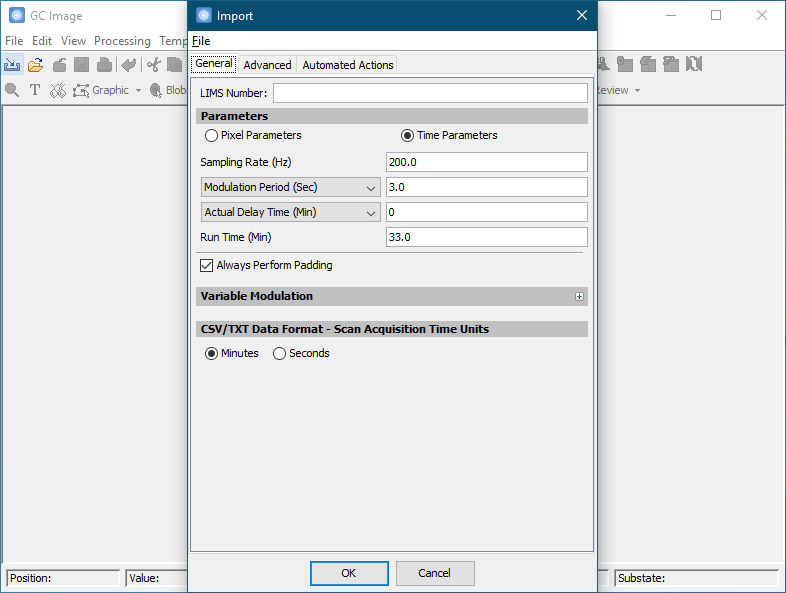
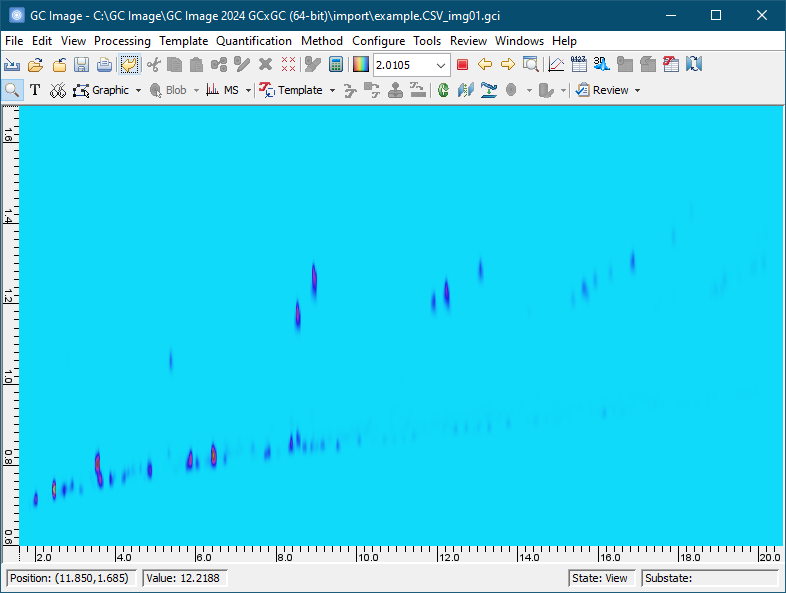
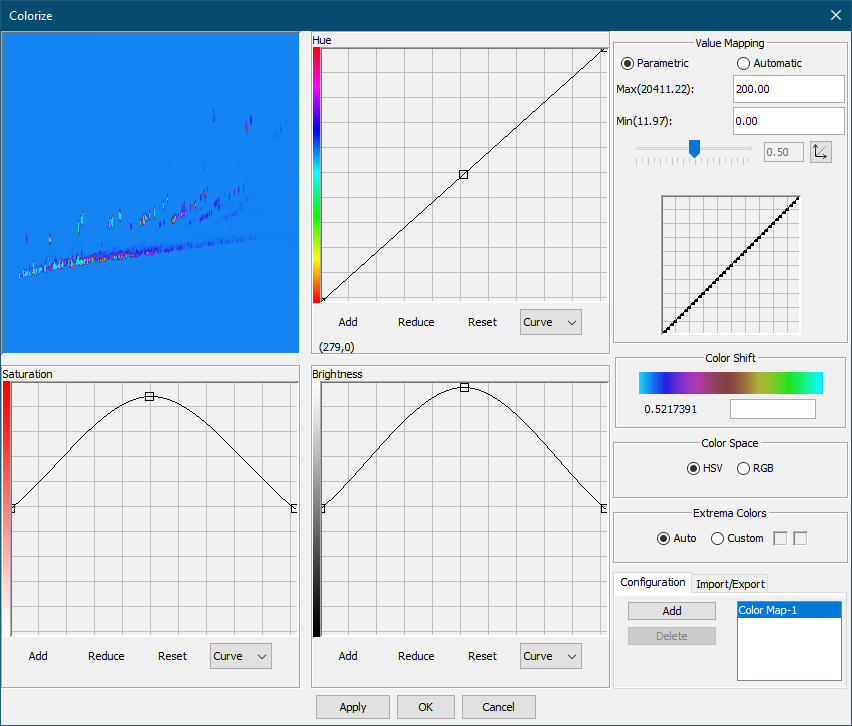
In the Colorize tool, set Max to 200 and Min to 0 under Value Mapping. Click OK to apply the new color map.
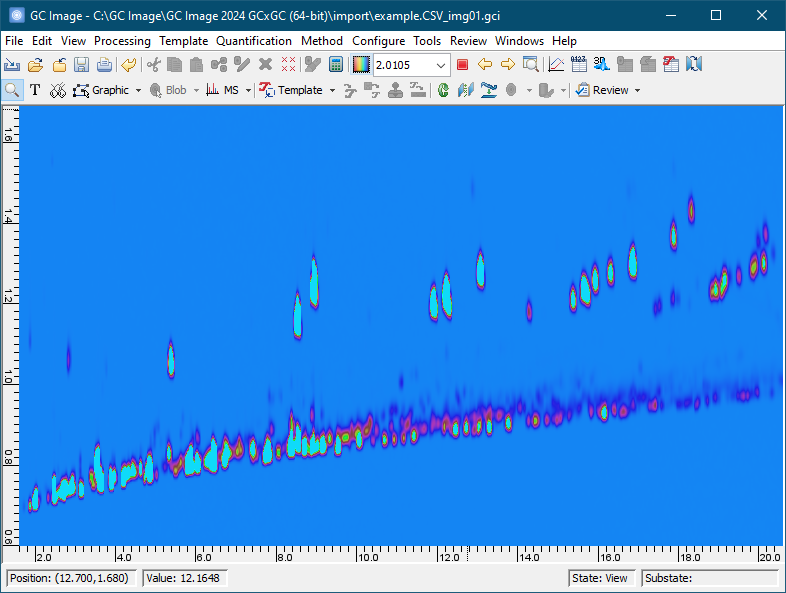
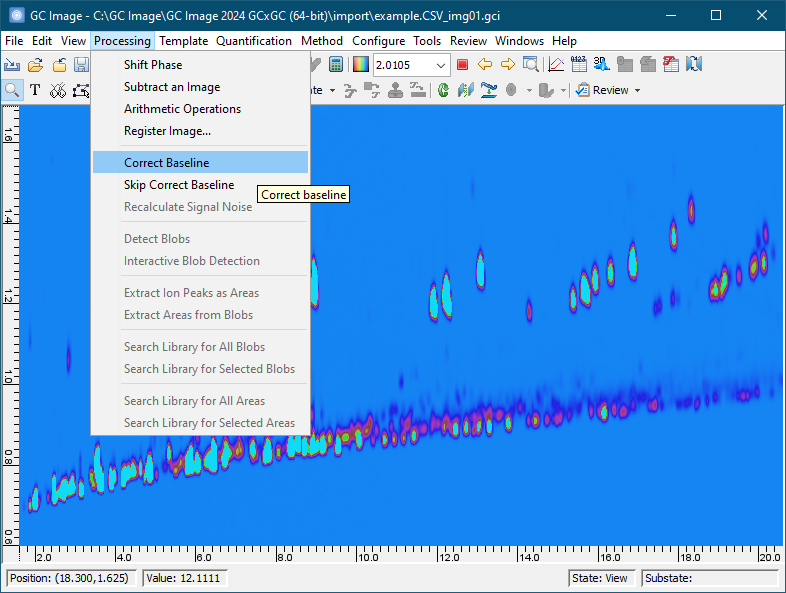
Click the Correct Baseline button on the tool bar (a tool tip is shown) or select Filter -> Correct Baseline from menu.
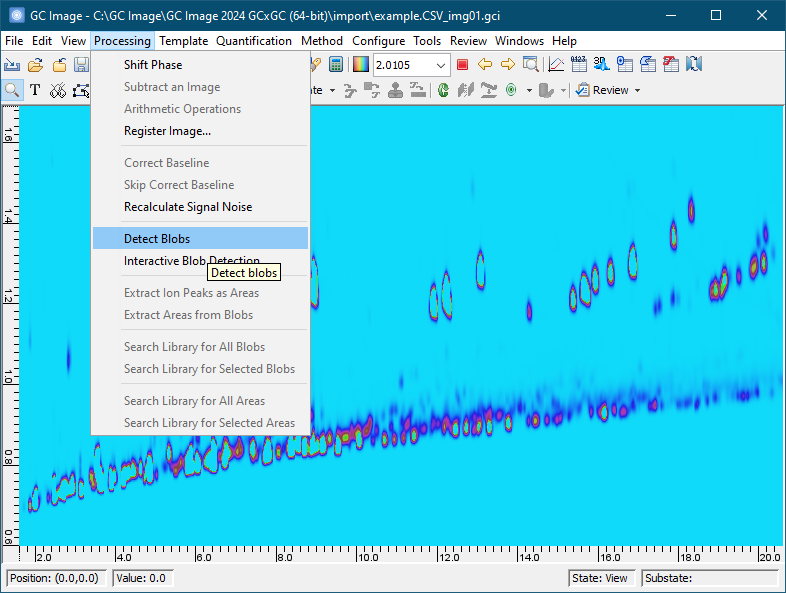
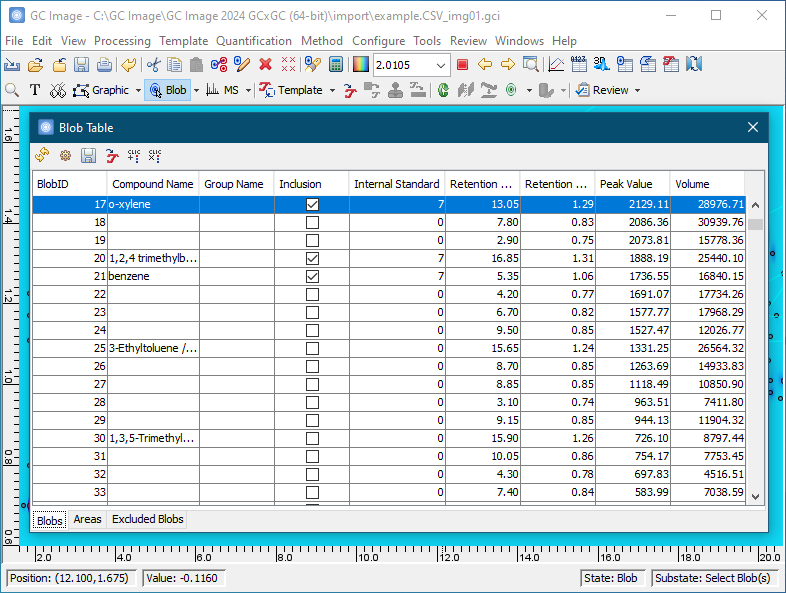
To select blobs for inclusion, first set the cursor mode on the Image Viewer palette to Blob Mode -> Select Blobs.
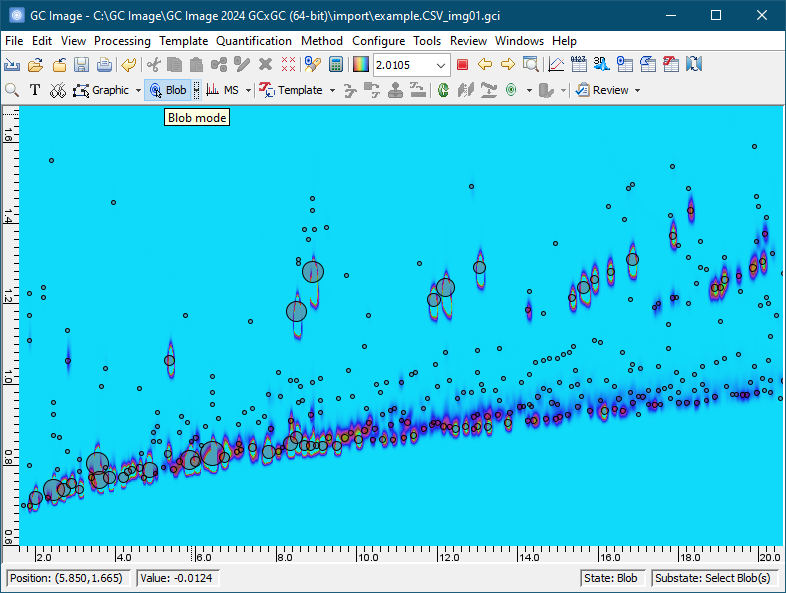
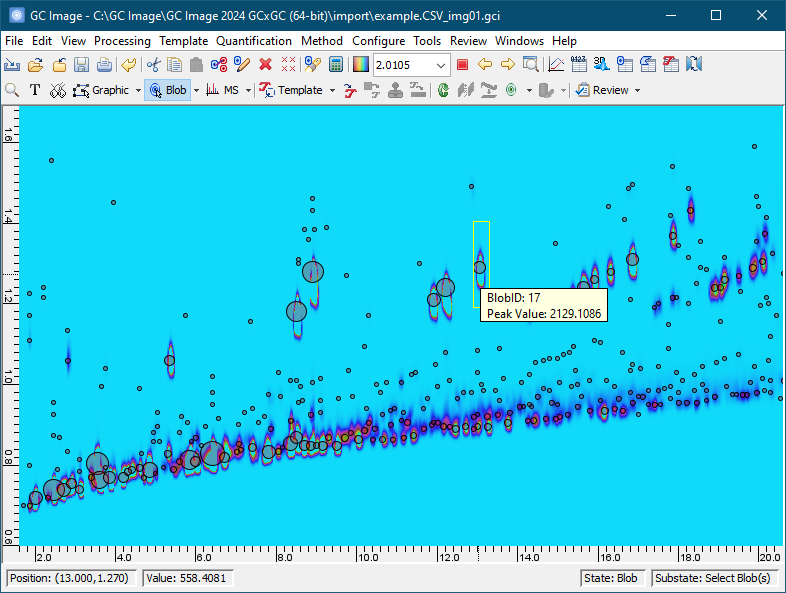
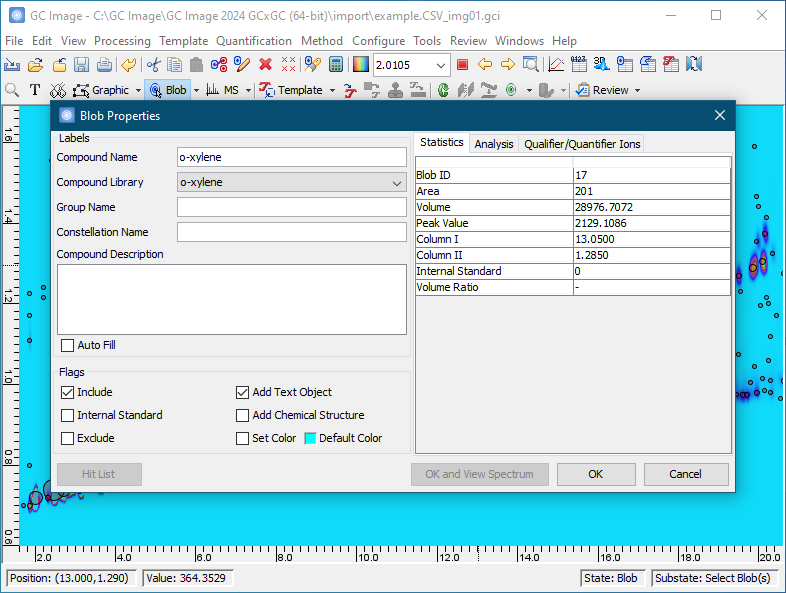
Subsequent chapters of the GC Image GCxGC Edition User Guide describe installation and details on using the software.
GC Image™ GCxGC Edition User Guide © 2001–2025 by GC Image, LLC, and the University of Nebraska.